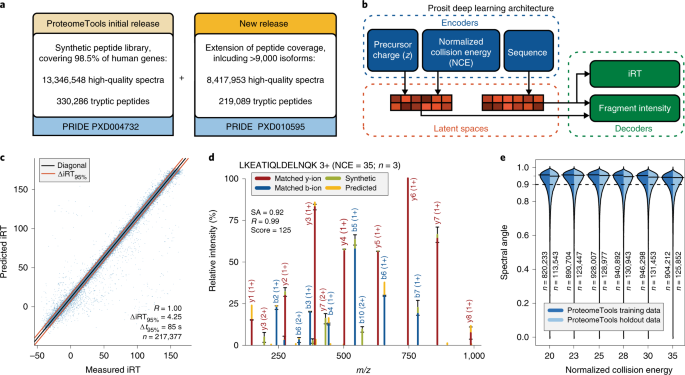
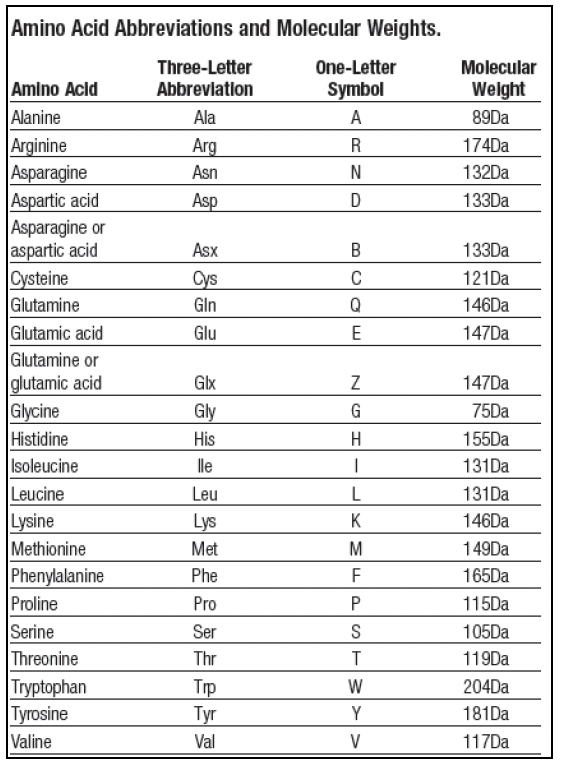
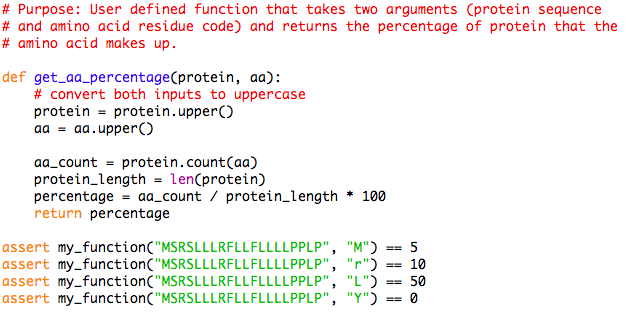
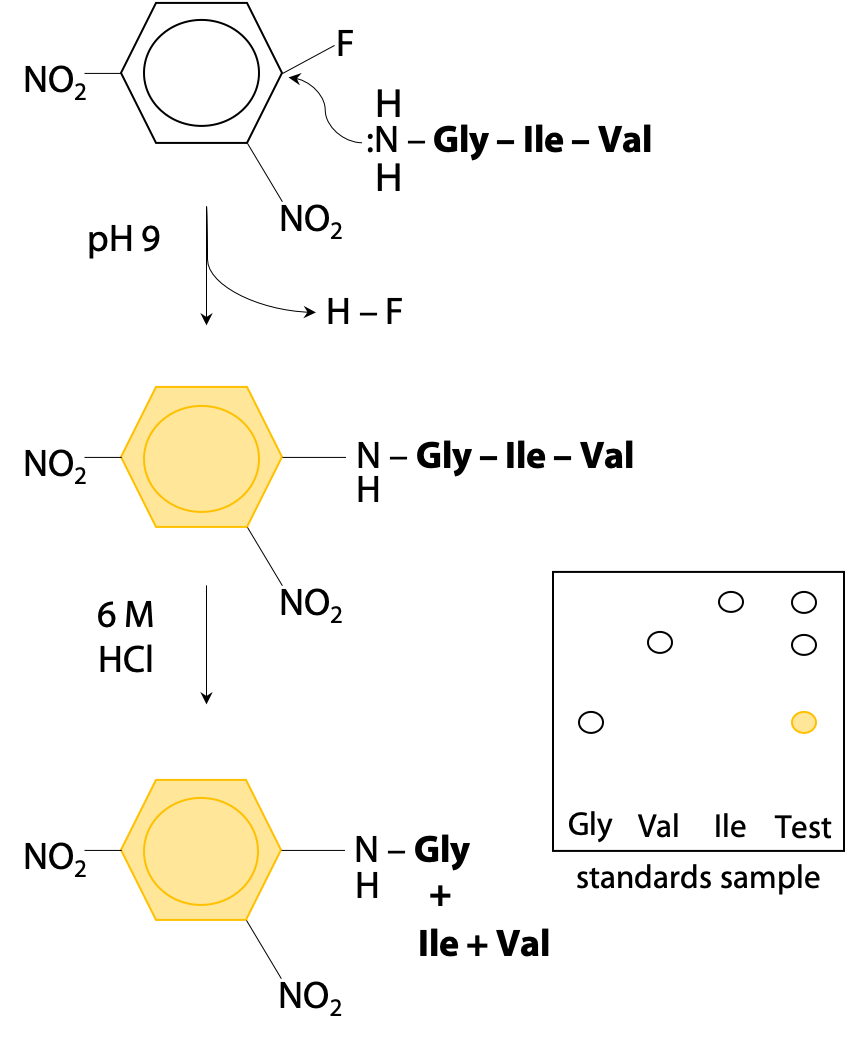
ProteoClade: A taxonomic toolkit for multi-species and metaproteomic analysis | PLOS Computational Biology
BioPython_Scripts/HowTo_Translate_DNA_fasta_to_Protein_Fasta.txt at master · PNNL-Comp-Mass-Spec/BioPython_Scripts · GitHub
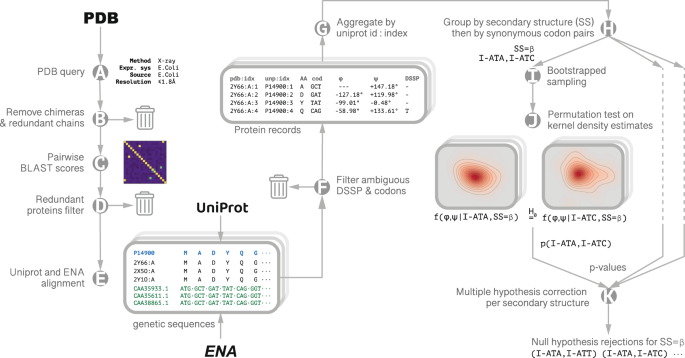
Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon | Nature Communications

Computational generation of proteins with predetermined three-dimensional shapes using ProteinSolver - ScienceDirect

Protein structure, amino acid composition and sequence determine proteome vulnerability to oxidation‐induced damage | The EMBO Journal

Data acquisition and database search. For a given experimental MS/MS... | Download Scientific Diagram
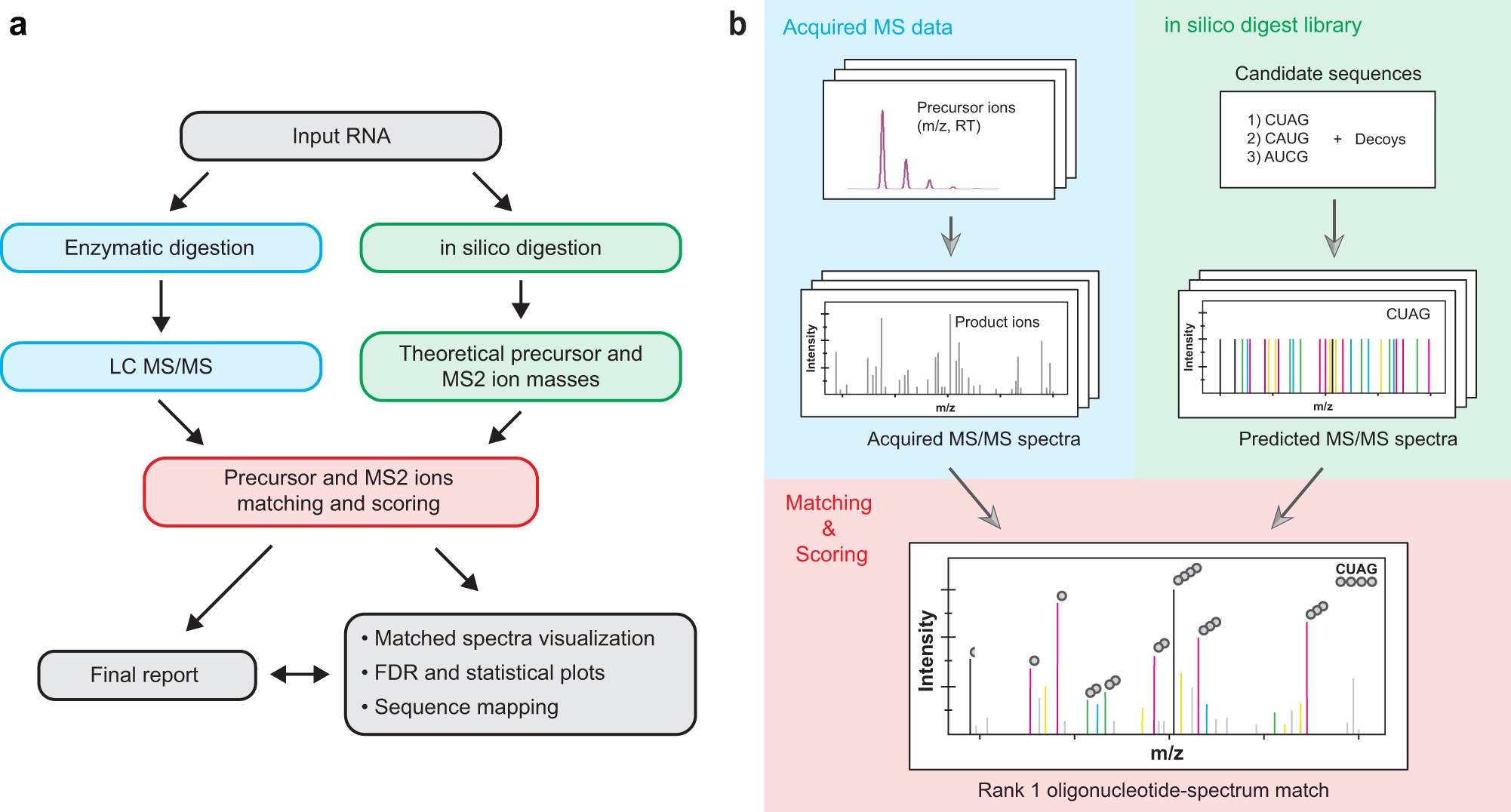
Pytheas: a software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry | Nature Communications
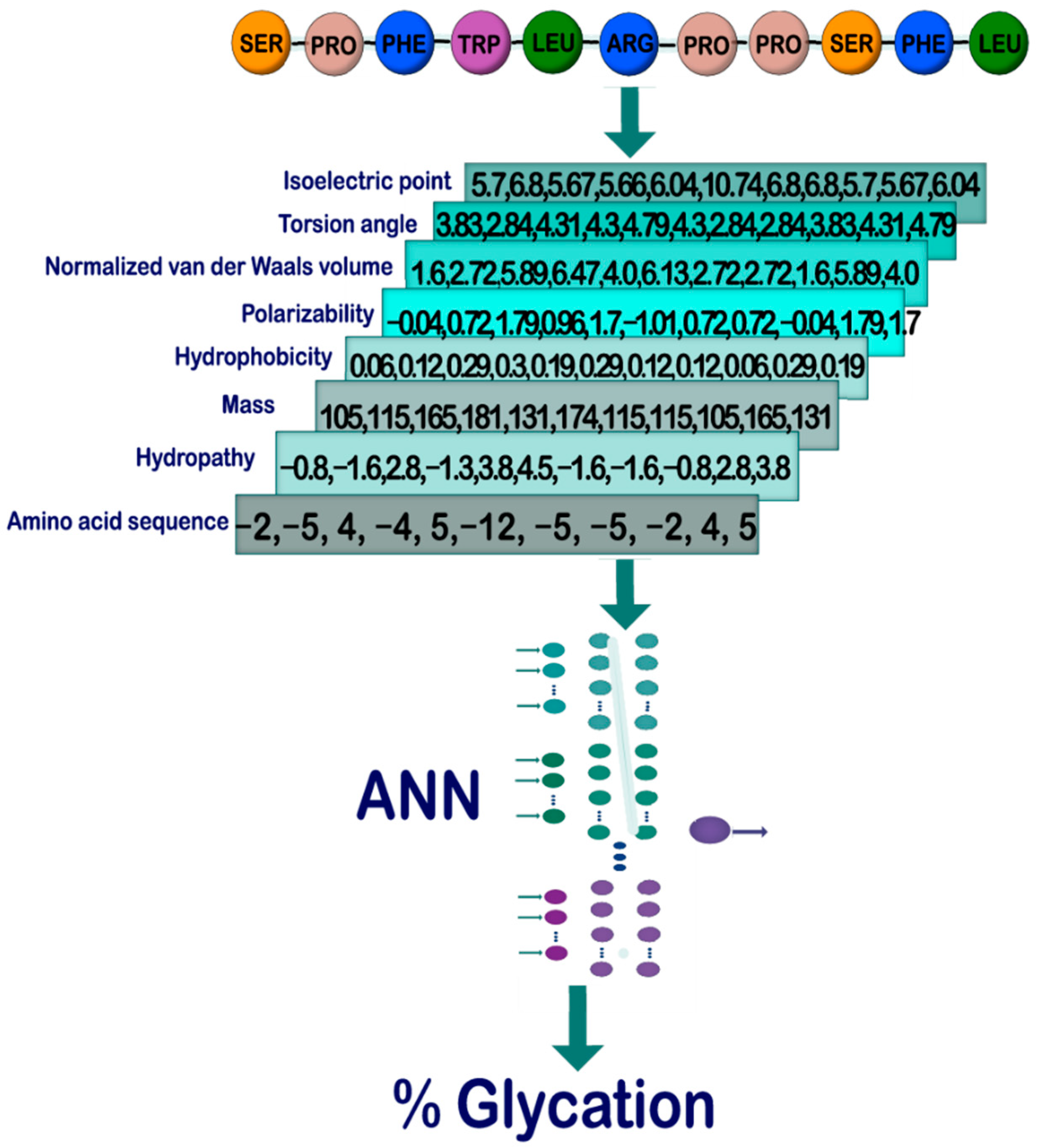
Sensors | Free Full-Text | On the Prediction of In Vitro Arginine Glycation of Short Peptides Using Artificial Neural Networks

Overview of the IPC 2.0 architecture. The input (amino acid sequence in... | Download Scientific Diagram